PubMed Central PMC01 is the U.S. National Institutes of Health's free digital archive of full-text biomedical and life sciences journal literature. Content is stored in XML at the article level. and is displayed dynamically from the archival XML each time that a user retrieves an article.
PubMed Central was started in 1999 to allow free full-text access to journal articles. Participation by journals is voluntary. From the beginning there has always been a requirement that participating journals provide their content to NCBI marked up in some "reasonable" SGML or XML format Beck01 along with the highest-resolution images available, PDF files (if available), and all supplementary material. Complete details on the PMC's file requirements are available PMC02.
The Promise of Marked-up Content
Building a full-text journal article repository seemed like a pretty straightforward task in when PMC was conceived in 2000. After all, participating journals already had their content in SGML or XML that they were putting online or sending to a vendor to be put online. All we would need to do would be to get the marked-up content, render it in PMC, and store it.
But we soon found out that the content we were receiving was not of the same quality as the articles that had been printed or even as the articles showing on the journals' websites. This seemed odd; the SGML was created for the online product, and this SGML was delivered to PMC for the archive. At the turn of the century, XML-first publishing workflows (where the articles are authored in XML or converted to XML as soon as they were submitted to a journal and then processed as XML) were not the norm.
The workflow that we encountered generally went something like this:
-
article is written, submitted, peer reviewed, revised, and accepted as a Word document (or more specifically, a printout of a Word document).
-
accepted article copyedited in the Word file (to capture the author's keystrokes).
-
copyedited Word file is ingested into a typesetting system and made into pages.
-
any changes in the author proof cycle are made in the typesetting system under the direction of copyeditors or proofreaders.
-
typesetting files are built into issues, checked by copyeditors or proofreaders and sent for printing.
-
typesetting files are then converted to SGML/XML - generally to a model defined by the online service.
-
SGML/XML files are converted to HTML, and the HTML is checked for errors.
-
HTML is corrected so that the online article represents the printed article.
-
SGML/XML files are stored away - never to be thought of again.
Things were going pretty well in that workflow up through item 5. The author's keystrokes had been used from the original word processing files—reducing errors that would have been introduced by rekeying the article; a person familiar with the content (usually at least two of a copyeditor, proofreader, or author) checked the files and any changes at each stage until it was sent off to press.
At this point it is out of the hands of the content folks and into the hands of the data converters. In step 6, the typesetting files used as the source for the transformation contained some information about the structure of the article, but it certainly was not enough to build a proper SGML/XML representation of the article. Assumptions are made during the transformation, and a surprising amount of hand tagging - copy and paste - was done to create the files. The problem areas are not surprising: metadata, tables, and math.
From the PMC point of view, the real tragedy in this (generalized and non-specific) workflow happens at step 8, when the files are reviewed by a proofreader but corrections are made to the output HTML files and not back in the source SGML files. Once we started processing those SGML files in the young PMC system, we took a close look at the output of our rendering of the files to identify any problems with the way our system was handling the files.
We started to find a good number of problems with the supplied SGML that were not in the HTML version available on the web - and a surprising number that were incorrect on the web. Unfortunately we did not count the errors that we encountered at this point. We simply fixed them, reported them to the publisher so that any that also appeared on their site could be fixed, and moved on the the next article or issue. This is when we knew we would have to have a team of content specialists checking all of the content we get in PMC.
PMC Ingest Workflow
The PMC processing model has been addressed in detail previously Beck01. Briefly it is diagrammed in Fig. 1. For
each article, we receive a set of files that includes the text in SGML or XML, the
highest
resolution figures available, a PDF file if one has been created for the article,
and any
supplementary material or supporting data. The text is converted to the current version
of the
NISO Archiving and Interchange Tag Set JATS01, and the
images are converted to a web-friendly format. The source SGML or XML, original images,
supplementary data files, PDFs, and NLM XML files are stored in the archive. Articles
are
rendered online using the NLM XML, PDFs, supplementary data files, and the web-friendly
images.
Fig. 1: PMC Processing Model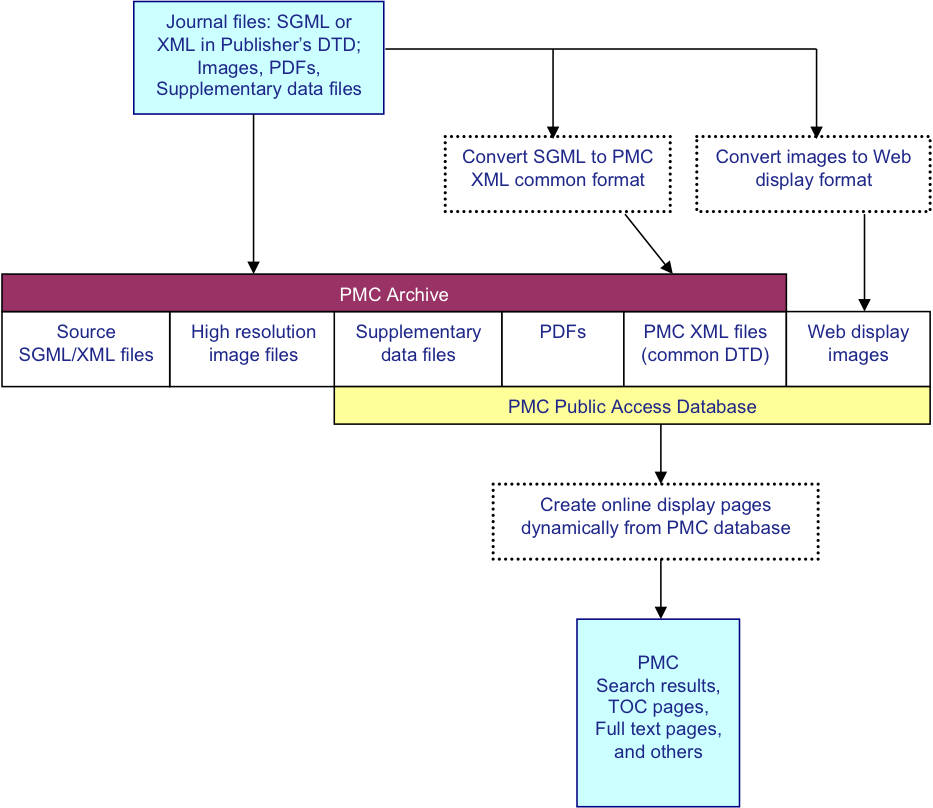
For purposes of this paper, we will concentrate on the text processing.
PMC Philosophy of Text Processing
There are four main principals to the PMC philosophy of text processing
First, we expect to receive marked-up versions of an article that are well-formed, valid, and accurately represents the article as it was published. We do not go so far as requiring that the content be true or correct as Syd Bauman illustrates Bauman01, merely that the article represents the version of record.
The question of what is the "Version of Record" is left up to the publisher. It may be a printed copy, a PDF version, or the journal's website.
Unlike the early days of PMC as described above, we do not correct articles or files. That is, we will not fix something that is wrong in the version of record nor will we make a correction to an XML file. All problems found in either processing or QA of files are reported back to the publisher to be corrected and resubmitted.
Thirdly, the goal of PMC is to represent the content of the article and not the formatting of the printed page, the PDF, or the journal's website.
And finally, we must run a Quality Assessment on content coming into PMC to ensure that the content in PMC accurately reflects the article as it was published. Our QA is a combination of automated checks and manual checking of articles. To help ensure that the content we are spending time ingesting to PMC is likely to be worthwhile, journals must pass through an evaluation process before they can send content to PMC in a regular production workflow.
The Data Evaluation Process
The data evaluation process has been described previously Beck01, but it is integral in ensuring the quality of content that is being submitted to PMC.
Journals joining PMC must pass two tests. First, the journal must pass PMC's scientific quality standard, which means that the journal must be approved for the NLM collection by NLM's Selection and Acquisitions section NLM01. This check ensures that the journal's content is "in scope" for a medical library and is of sufficient scientific quality for the archive.
Next the journal must go through a technical evaluation to "be sure that the journal can routinely supply files of sufficient quality to generate complete and accurate articles online without the need for human action to correct errors or omissions in the data." PMC03
For the data evaluation, a journal supplies a set of sample files for approximately 50 articles. These files are put through a series of automated and human checks to ensure that the XML is valid, accurately reflects the publication model of the journal (publication dates, for example) and is a complete and accurate representation of the published articled. There is a baseline set of "Minimum Data Requirements" that must be met before the evaluation proceeds to the more human-intense content accuracy checking PMC04. These minimum criteria are listed briefly below:
-
Each sample package must be complete: all required data files (XML/SGML, PDF if available, image files, supplementary data files) for every article in the package must be present and named correctly.
-
All XML files must conform to an acceptable journal article DTD.
-
All XML/SGML files must parse according to their DTD.
-
Regardless of the XML/SGML DTD used, the following metadata information must be present and tagged with correct values in every sample file:
-
Journal ISSN or other unique Journal ID
-
Journal Publisher
-
Copyright statement (if applicable)
-
License statement (if applicable)
-
Volume number
-
Issue number (if applicable)
-
Pagination/article sequence number
-
Issue-based or Article-based publication dates. Articles submitted to PMC must contain publication dates that accurately reflect the journal’s publication model.
-
-
All image files for figures must be legible, and submitted in high-resolution TIFF or EPS format, according to the PMC Image File Requirements.
These seem like simple and obvious things - xml files must be valid - but the minimum data requirements have greatly reduced the amount of rework that the PMC Data Evaluation group has to do. Certainly it helps to be explicit about even the most obvious of things.
During the data evaluation, PMC reviews 100% of the sample articles by eye.
Production QA
The PMC production team is made up of a set of Journal Managers who are responsible for the processing and checking of content for the journal titles that are assigned to them. We use a combination of automated processing checks and manual checking of articles to help ensure the accuracy of the content in PMC.
We can leverage the fact that all content is sent to us in SGML or XML to eliminate any article files that are not well-formed (if XML) or valid to the model they claim to be valid against. 100% of not-well-formed or invalid files are returned to the provider to be corrected and resubmitted.
The PMC Style Checker (Beck01) is used during the ingest process to ensure that all content flowing into PMC is in the PMC common XML format for loading to the database. The errors provided by the Style Checker provides us with a level of automated checking on the content itself that can highlight problems, but it only goes so far. For example, the Style Checker can tell you if an electronic publication date is tagged completely to PMC Style (contains values in year, month, and day elements) in a file, but it can't tell you if the values themselves are correct and actually represent the electronic publication date of the article.
PMC also has a series of automated data integrity checks that run once content is loaded to the database which can identify, among other things, problems like duplicate articles submitted to the system, and potential discrepancies in issue publication dates for a group of articles in the same issue.
Early in the PMC project the questions of "what and how much to check?" were left to the discretion of the Journal Manager. But we soon found out that certain things had to be checked more frequently. All articles with marked-up math (in MathML or LateX) get a close review because the quality of marked up math has been one of the problem areas since the beginning of the project. In addition, all published corrections and retractions needed to be checked closely to ensure that the tagging provided in the source XML allows PMC to build a link between the correction/retrction and the article being corrected/retracted. We also check the ever PDF that we generate for manuscripts being tagged for the NIH Public Access Project NIH01.
As the project began to grow, both in terms of the amount of content coming into PMC and the number of Journal Managers required to manage the content, it became obvious that we needed to quantify the QA process. First, the production team compiled a "Content QA checklist" which was a collection of some of the most common (and serious) errors encountered over the years. Next, we built a system that shows each Journal Manager the journals that are assigned to him, and which articles need to be checked Fig. 2. The system selects a percentage of articles from each "batch" of new content deposited by the journal as they are processed and loaded to the PMC database. The number of articles in each batch is determined by the publication model and participation mode of the journal itself. Traditional journals which deposit a whole issue at a time into PMC will normally have a "batch" consisting of all the articles in the issue. Journals which publishing on a continuous basis and send content to PMC article-by-article will have batches which reflect how much content is deposited during a given day.
While it would be nice to be able to check 100% of the articles being loaded into
the PMC
database, this is generally not reasonable because of resources. So, once an journal
has
passed the tightly monitored data evaluation period, and has demonstrated over a period
of
time after moving into production that it can provide generally error-free data, QA
is done on
more of a spot check basis. By default, new journals that come out of data evaluation
and move
into production are set with a higher threshold of articles selected for QA in the
system.
Once PMC's Journal Manager is confident in the journal's ability to provide good,
clean, data,
the article-selection percentages are lowered over time. Journals with a proven track
record
of providing error-free data into PMC generally have lower percentage of articles
checked for
QA. If the Journal Manager begins to see problems on an ongoing basis, the percentage
of
articles checked may be increased.
Fig. 2: QA System Dashboard
The system includes the original list of standard errors that was compiled as the
first
"Content QA checklist"; this list has grown the years as new errors are encountered.
PMC
Article Errors are grouped into eight major categories: Article Information, Article
Body,
Back Matter, Figures and Tables, Special Characters and Math, Generic Errors, Image
Quality,
and PDF Quality. Within each of these major categories, there may be one or more
sub-categories. For example, in the "Article Body" section there is a subcategory
for
"Sections and Subsections", containing errors for mising sections, or sections that
have been
nested incorrectly in the flow of the body text. In Fig. 3 the errors specific
to Article Information are shown in the purple box. If a Journal Manager finds that
the
"Publication date is missing or does not match the version of record", then he or
she simply
checks that box and that error is recorded for that article. Of course, there is always
a text
box available to allow a Journal Manger to enter an unforeseen error.
Fig. 3: Article Errors List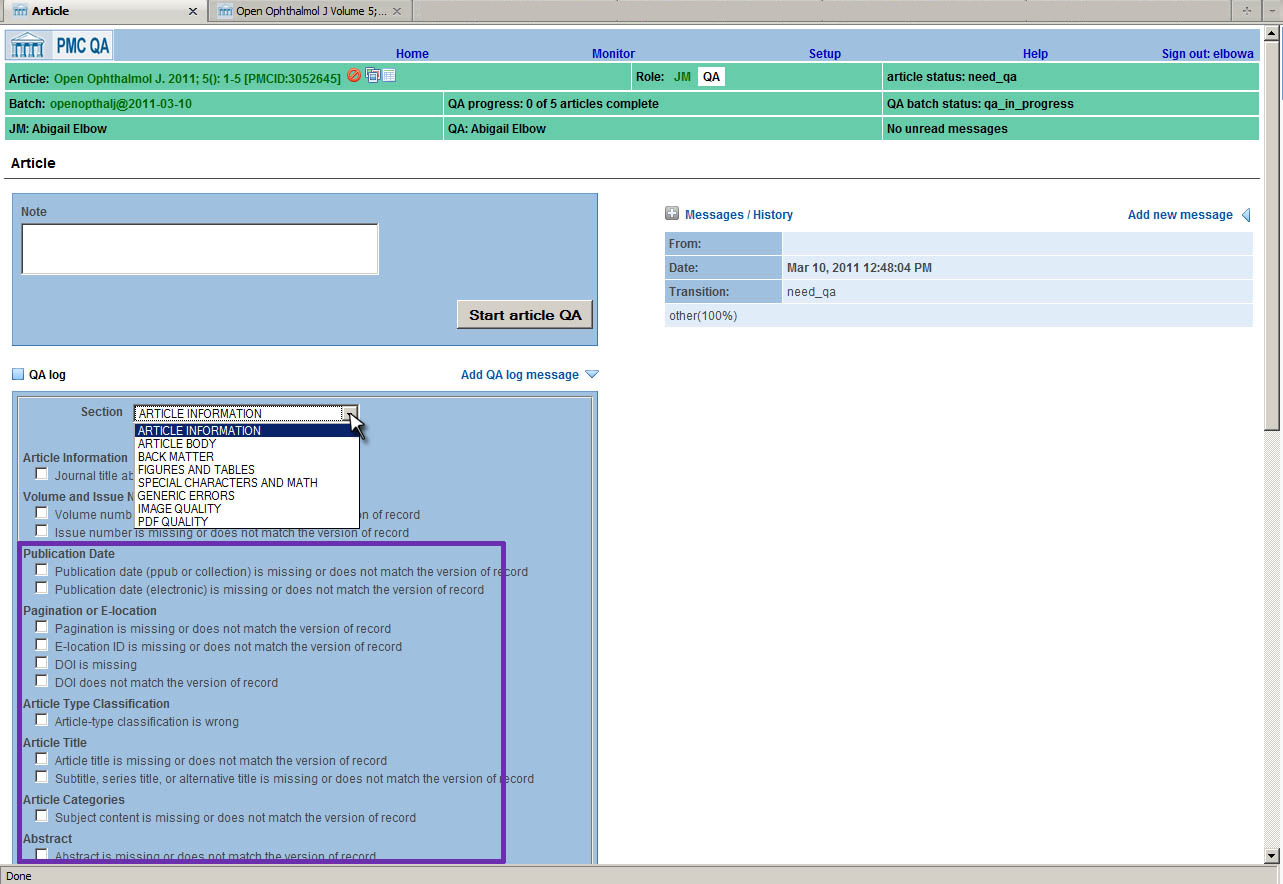
Processing can be done either on the issue level or on a group of articles that arrives
at PMC in a given time period. Each error is classified as "severe" or "normal" and
as a "PMC
Error" or a "Data Error". See the purple box in Fig. 4 for an example of a
totaled batch of articles.
Fig. 4: Error Totals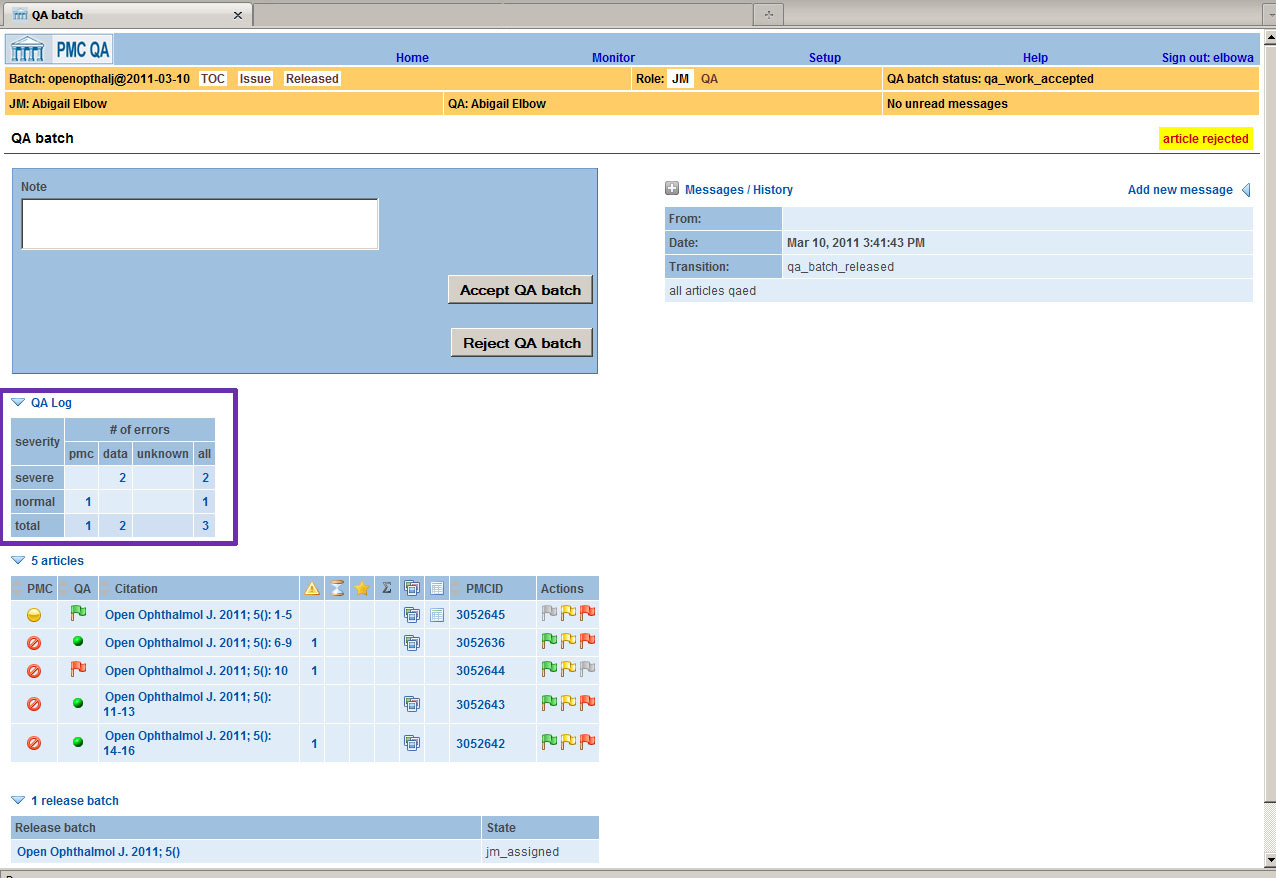
At any time, the Journal Manager can review the accumulated errors for a batch as
seen in
Fig. 5. This report can be output in RTF format so it can be sent to the
publisher as a Word file (Fig. 6>).
Fig. 5: Batch Errors Fig. 6: Batch Error Report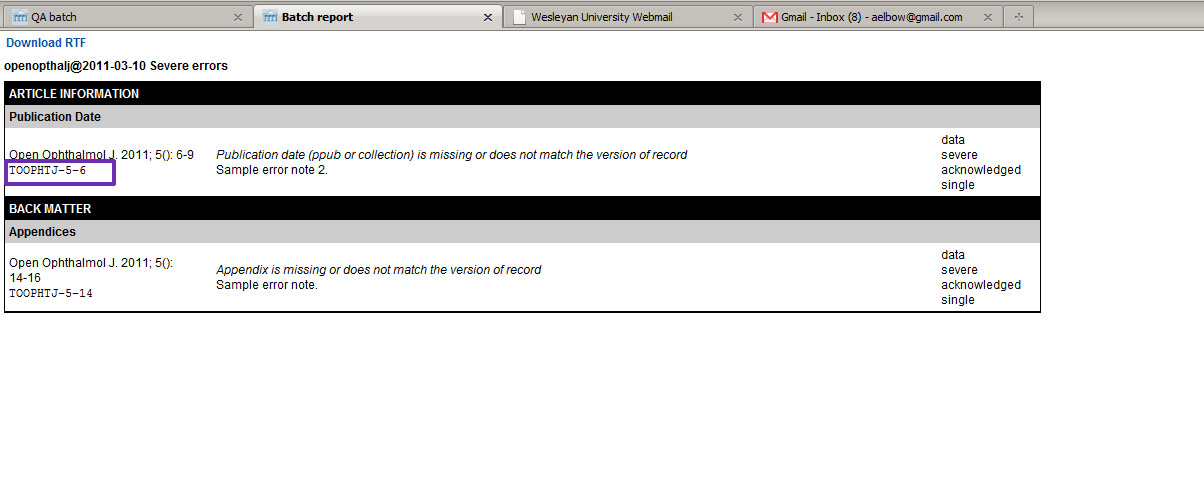

Conclusions
The automated QA system has greatly improved both the Quantity and the Quality of the QA work that is being done for the PMC project, but we find that even this long after the 10-year anniversary of the invention of XML that there is still a need for manual review of article files.
Fortunately we are able to sort out the deepest structural problems with XML tools, but questions of accuracy of content still need to be addressed by eye.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Library of Medicine, National Center for Biotechnology Information.
References
[PMC01] PubMed Central, http://www.ncbi.nlm.nih.gov/pmc/.
[Beck01] Beck, Jeff. “Report from the Field: PubMed Central, an XML-based Archive of Life Sciences Journal Articles.” Presented at International Symposium on XML for the Long Haul: Issues in the Long-term Preservation of XML, Montréal, Canada, August 2, 2010. In Proceedings of the International Symposium on XML for the Long Haul: Issues in the Long-term Preservation of XML. Balisage Series on Markup Technologies, vol. 6 (2010). doi:https://doi.org/10.4242/BalisageVol6.Beck01.
[PMC02] How to Join PMC, http://www.ncbi.nlm.nih.gov/pmc/about/pubinfo.html.
[JATS01] NISO Journal Article Tag Suite (JATS), http://jats.nlm.nih.gov/archiving/.
[Bauman01] Bauman, Syd. (2010) "The 4 Levels of XML Rectitude", Balisage 2010, poster.
[NLM01] NLM Collection Development and Acquisitions, http://www.nlm.nih.gov/tsd/acquisions/mainpage.html.
[PMC04] Minimum Criteria for PMC Data Evaluation Submissions, http://www.ncbi.nlm.nih.gov/pmc/pmcdoc/mindatareq.pdf.
[PMC03] How to Join PMC, http://www.ncbi.nlm.nih.gov/pmc/about/pubinfo.html.
[NIH01] National Institutes of Health Public Access http://publicaccess.nih.gov/