Introduction
The STAR (Self-defining Text Archival and Retrieval) file format for electronic data transfer and archiving was introduced in 1991 [ Hall, 1991 ], many years before the introduction / standardization of XML. The STAR format was designed to be extensible and flexible to handle all types of data in a machine independent manner. To achieve these goals, the STAR format is text-based using domain-specific keys associated with values. Importantly, STAR attempts to provide an easy manner for representing and retrieving tabular data across multiple datasets. In that context, additional considerations are that the data definitions for tables which are unique within a single dataset are also reusable across datasets. These concepts will be discussed in greater detail as the STAR model is deconstructed.
The STAR file format has been defined through a series of manuscripts published from 1991 through 2012 [ Hall, 1991; Hall & Spadaccini, 1994; Spadaccini & Hall, 2012 ]. Through these series of papers the STAR format has been detailed, refined, extended and further embellished with specifications for a Dictionary Definition Language [ Hall & Cook, 1995 ] – a construct analogous to an XML Schema Definition or its predecessor, the Document Type Definition.
While STAR predates XML, it has not been adopted by as extensive a user-base as XML. That said, STAR is the primary data format used by several important subfields of biochemistry, including structural biology using either x-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, or cryo-electron microscopy. STAR is the file format in use at the Protein Data Bank (PDB) and Biological Magnetic Resonance Bank (BMRB) for both data archival and retrieval as well as for the maintenance and development of data dictionaries for these scientific disciplines (for instance for standardizing the Molecular Information File (MIF) [ Allen, et al., 1995 ], the Crystallographic Information File (CIF) [ Hall, et al., 1991 ] and the NMR-STAR [ Ulrich, et al., 2008 ] dictionaries).
Manuscript Outline
The goal of this manuscript is to deconstruct the structure from the syntax of the STAR format. This will entail a broad description of the core data structures implicit to STAR, followed by a codification using the OCaml programming language, followed by eventual transcoding of the structures into a proposed core XML schema definition. OCaml is a functional programming language; however, the reason for choosing OCaml for this portion of the work is that OCaml is a statically-typed programming language relying on type inference. This makes it well-suited for this task and for coding a generic STAR parser and translator to XML. This code is available on Github and examples of its use are given at the end of this manuscript.
Data Model
There are five basic data structures defined in STAR. There are three container constructs [1] which define the scope of the data; the other two structures allow for either tabular data or data which are recorded as key-value pairs. As STAR is a file format, there is also a fourth implied container structure which is the file itself. A contrived example of a STAR file is provided in Appendix A to help illustrate these structures.
Containers
The various container constructs in STAR define the scope of the uniqueness of the data names. Within any container, data names are required to be unique, that is, used only once. This helps support efficient data retrieval from the file.
The top level container is the file itself. It is implicit that all of the data of interest are stored within the STAR file, and if there are other versions of the same file there is no guarantee of uniqueness between the files. Reiterating that STAR is a file format, the file is the entire domain for the STAR specification.
There are two file-level containers allowed within a STAR file, data blocks and global
blocks. A STAR file must contain at least one data block to be valid. Data blocks
carry unique names within the file; in the STAR syntax they are defined with the
prefix data_ such as data_blockname. There is no
significance to the order of data blocks within the file, the only constraint being
the uniqueness of the name.
The second file-level container is a global block. Unlike data blocks, global blocks are unnamed. They rely on document order as a means to maintain state information throughout the file. The interpretation is that if several global blocks contain the same data items, those items are "over-written" with subsequent global blocks [ Hall & Spadaccini, 1994 ] . This may lead to multiple, ambiguous interpretations if the global blocks contain identical table structures; however, this manuscript is focused on structure and syntax, not the semantics of global blocks.
Both data blocks and global blocks may contain either tabular data (loops [2] ) or key-value pairs (items). Within any block the data names must be unique. Data blocks may also contain an additional container item, the save frame. Save frames are similar to data blocks in that they are given unique names within a data block, and save frames may contain loops or data items. According to the 1994 specification, save frames cannot contain other save frames. The 2012 specification allows nested save frames allowing for a limitless depth hierarchy. This is shown below as a possible XML instance.
Figure 1: An example of the hierarchical structure of the STAR containers.
<STAR-file>
<data name="block_1">...</data>
<global>...</global>
<data name="block_2">
<save name="frame_1">
<save name="frame_1.1">...</save>
</save>
</data>
<data name="block_3">...</data>
<global>...</global>
<data name="block_4">...</data>
<global>...</global>
</STAR-file>One final note about containers: they must contain some data, either a table or a key-value pair as described in the following section.
Basic Data Structures
There are two basic data structures allowed in STAR. "Data items" are key-value pairs representing a single data value associated with an element. "Loops" are for representing tabular data where there are one or more data records containing one or more data values [3]. Data names must be unique within the scope of any container; the name for a key cannot be used for a column header within the same data block, global block or save frame.
Key Value Pairs
Key value pairs in the core STAR standard are simple and straightforward. Keys and values are separated by whitespace. Keys (also referred to as data names) begin with an underscore. Keys have no character constraints other than that they cannot contain whitespace. This makes it challenging to map keys as XML element names, as in practice the keys can contain ‘[‘ and ‘]’ characters [4]. Data values are always treated as the equivalent of strings regardless of whether the value represents an integer, a float, a Boolean or any other basic data type [5]. The one exception to this is that STAR supports listing a frame_code as a value, where frame_code is the name of a save_frame contained within the corresponding data block. There are various conventions for reporting NULL values, a ‘?’ in mmCIF or a ‘.’ in NMR-STAR.
There are four different ways to delimit ordinary values. White space delimited values cannot contain white space. Single or double-quoted values can contain spaces and tabs but must be contained on a single line. Semi-colons are used to delimit multi-line values. An example XML instance is:
<datum name="location" type="string" delimiter="space">California</datum>
Tabular Data
A simple, non-nested table is defined syntactically with the keyword
loop_.
This is then followed by a white-space separated list of data names (keys or column
headers) and a list of data values (which can be delimited in
any manner). The specification demands that in the case of a simple table, the
number of data values must be an integral multiple of the number of keys, that is,
every cell in the table is given a value. As mentioned above, there are several
conventions for reporting NULL values. In core STAR, there is no syntactical keyword
defining the end of a table. A table ends when no more values are listed, that is,
when another table, key, or container is found, or the file ends. However, in
NMR-STAR, tables are formally terminated with the keyword stop_.
STAR also provides for nested tables. Nested tables are both fascinating and powerful
as they represent a type of relational structure. The syntax of nesting tables
is simply to insert another loop_ keyword within the list of column
headers. The list of values will then be interpreted as filling up one full row of
the table followed by rows in the smaller, nested table until encountering a
stop_ keyword. stop_ signifies that the following values
belong to the closest outer table.
It is easiest to demonstrate this complicated structure using the examples provided in Hall & Spadaccini, 1994 shown below. (The keys and values used in these examples are significant to chemists but for the purposes of this manuscript, they can be treated as arbitrary names.)
loop_
_atom_identity_node
_atom_identity_symbol
loop_
_atom_bond_node_1
_atom_bond_node_2
_atom_bond_order
A1 B1 1 2 single stop_
A2 B2 1 6 double 30 40 triple stop_
A3 B3 1 7 single stop_
Table I
Illustration of the interpreted layout of nested tables
| atom_identity_node | atom_identity_symbol | atom_bond_node_1 | atom_bond_node_2 | atom_bond_order |
|---|---|---|---|---|
| A1 | B1 | 1 | 2 | single |
| A2 | B2 | 1 | 6 | double |
| 30 | 40 | triple | ||
| A3 | B3 | 1 | 7 | single |
In this example, the outer table contains the “atom identity” columns while
the inner one contains the “atom bond” information. The first five values become
row A1; the stop_ keyword signifies that the next values start back at
the outer table. Similarly the next five values become row A2. However, the values
30, 40, triple become a second row in the nested table. The stop_ keyword signifies
the remaining five values belong to the third row of the outer table.
This nested structure which is easiest to visualize using an XHTML table as shown above, can be interpreted as a relational structure in which two entities (the atom_identity entity and the atom_bond entity) are related by an implicit foreign key. In this manner, the node pair of A2 and B2 can have two atom bond records associated with it while the others only have one.
A more complicated double nesting from Hall & Spadaccini, 1994 is shown below.
loop_ _atomic_name loop_ _scheme _atomic_energy loop_ _function_exponent _function_coefficient hydrogen (2)->[2] -0.485813 1.3324838E+01 1.0 2.0152720E-01 1.0 stop_ (2)->[2] -0.485813 1.3326990E+01 1.0 2.0154600E-01 1.0 stop_ (2)->[1] -0.485813 1.3324800E-01 2.7440850E-01 2.0152870E-01 8.2122540E-01 stop_ (3)->[2] -0.496979 4.5018000E+00 1.5628500E-01 6.8144400E-01 9.0469100E-01 1.5139800E-01 1.0000000E+01 stop_ stop_
Table II
Illustration of the tabular layout of double-nested loops
| atomic_name | scheme | atomic_energy | function_exponent | function_coeffecient |
|---|---|---|---|---|
| hydrogen | (2)->[2] | -0.485813 | 1.3324838E+01 | 1.0 |
| 2.0152720E-01 | 1.0 | |||
| (2)->[2] | -0.485813 | 1.3321990E+01 | 1.0 | |
| 2.0154600E-01 | 1.0 | |||
| (2)->[1] | -0.485813 | 1.3324800E-01 | 2.7440850E-01 | |
| 2.0152870E-01 | 8.2122540E-01 | |||
| (3)->[2] | -0.496979 | 4.5018000E+00 | 1.5628500E-01 | |
| 6.8144400E-01 | 9.0469100E-01 | |||
| 1.5139800E-01 | 1.0000000E+01 |
This again points to the relational-like structure for the nested table with implicit primary/foreign keys. Note that this cannot be accommodated simply by replicating the left-most field among all nine rows; there are two distinct (2)->[2] records which both share the identical attribute name.
The table shown above is encoded using the Docbook version of an XHTML table. It is an obvious choice for an XML schema for STAR nested loops. However, one of the goals of this exercise is to maintain document order when converting between STAR and XML. For that reason, an alternate schema has been designed, an instance of which is shown below. It is an ongoing task to design an XSLT which can convert from this schema to the XHTML table schema.
<loop>
<header>
<column key=“column_1"/>
<header>
<column key=“column_2"/>
</header>
</header>
<row>
<cell>A1</cell>
<rows>
<row>
<cell>B1</cell>
</row>
</rows>
</row>
<row>
<cell>A2</cell>
<rows>
<row>
<cell>B2</cell>
</row>
<row>
<cell>B3</cell>
</row>
</rows>
</row>
</loop>Whitespace and Comments
Whitespace in the early STAR papers is defined as spaces, tabs and newlines. The use of whitespace is arbitrary, in that key value pairs can be stored within one line of text (separated by spaces and/or tabs) or they can span multiple lines of text. Similarly, whitespace can be used to create a tabular appearance for tabular data within a loop structure, but it is not required to. The only rules regarding whitespace is that if it appears within a data value than the data value must be delimited by single-quotes, double-quotes or semi-colons; and if the data value is multi-line it must be delimited by semi-colons.
Comments within STAR begin with a '#' symbol and continue until the end of the line. Comments have no meaning within a STAR file; they are considered to be the same as whitespace. Despite this formal specification, comments are important operationally to users and therefore it is a design consideration to translate comments from STAR to XML as well. This is an important benefit of XML over another possible serialization format, JSON, as JSON does not have support for comments. Not only are comments important to users, the positioning of comments is also important. This presents an interesting problem of maintaining document order of the comments when they can appear at almost any location within a STAR file. That is, a comment might be on the first line and be at the File container level of the hierarchy; conversely, a comment might appear between a key and a value or between values in a loop. These comments need to be mapped within the XML elements in which they appear. Such mapping of comments within the XML schema is still work in-progress.
There are three basic options for how to deal with comments in the STAR file. The first is to follow the STAR specification and treat them as whitespace which is to be ignored. This has a drawback in that it is more challenging to roundtrip a file conversion from STAR to XML and back to STAR again and verify that the conversion was lossless. However, that is a general problem with whitespace and the traditional solution is to accept canonical whitespace interpretations. However, ignoring comments also has a second drawback in that comments provide important information to real-world users. For instance, the ambiguity codes for NMR-STAR chemical shifts are described within a comment section of the STAR file. While the specification may state that STAR comments are to be treated as whitespace, comments are of value to the communities which use STAR.
A second option is to accept that comments are important for the file but to acknowledge that since there is no specification to their structure within the file, all comments are File level elements. That is to say, comments would not be attributable to data blocks, tables or key-value pairs, only the file itself. This would allow for a simple way of retaining the comments through translation; however, it would not allow the preservation of document order for the comments. Once plucked to the level of the file, it would not be possible to restore their original position without some other method of recording their position.
The third option is to design a grammar which associates comments with whichever data structure (XML element) they are within close proximity to. Note that the nature of comments means that such an association could never be perfect in terms of the abstract data structures; however, it would allow for preservation of document order. The example STAR file in Appendix B illustrates the complexity and inherent ambiguity of computationally interpreting the context of a given comment.
OCaml
As mentioned in the outline section, after deconstructing syntax from structure, the next goal was to codify the structure in OCaml and build an OCaml to XML translator. OCaml was chosen due to its strong static typing and type inference system. This makes OCaml particularly suited for this task.
Figure 2: OCaml STAR data structures
type key = string
type value = Space of string | Quote of string | Apos of string | Semi of string | Frame of string
type datum = key * value
type header = Key of key | Header of header list
type framecode = string
type datacode = string
type cell = Cell of value | Rows of row list
and row = Row of cell list
type loop = (header list) * (row list)
type block_items = Loop of loop | Datum of datum | Save of save_frame | Comment of string
and save_frame = framecode * (block_items list)
(* this is more permissive than specification. Can't have save_frame in global *)
type global_block = block_items list
type data_block = datacode * (block_items list)
type star_contents = Global of global_block | DataBlock of data_block | Comment of string
type star_file = star_contents list
A brief synopsis of OCaml data types, the built-in data type used is string.
New data types can be defined and variants are created using constructors which begin
with
an uppercase letter (ex. Quote or Apos). Finally, OCaml also has built-in data types
for lists and pairs, the latter defined using the '*'.
OCaml also supports recursive data types which are part of the STAR specification.
The
keyword and defines the nested tabular data as cells as well
as the nested save frames.
OCaml provides modules for lexing and parsing from a grammar. Decomposing STAR structure
from syntax allowed for a simple coding of both a lexer and parser for STAR files,
mapping all of the data into pairs and loops within the nested
container structures. Translating to XML then became a simple matter of traversing
this parse tree and serializing the data as XML. All of the OCaml code including the
XML schema developed for STAR is available on GitHub.
Deliverables
The exercise of deconstructing STAR syntax from structure is useful in understanding
the STAR specification as well as identifying versions and varients which are either
in the literature or currently in use. (For example, the additional stop_
keyword required by NMR-STAR). It is also useful in that it provides many concrete
deliverables which are available on GitHub as well as through other sources.
-
STAR XSD: An XML schema definition has been developed which attempts to be faithful to the STAR structure.
-
STAR2XML: OCaml code for translating STAR to the XML schema has been developed. A screenshot of this STAR-XML is shown below:
Figure 3: Screenshot of a Portion of STAR-XML
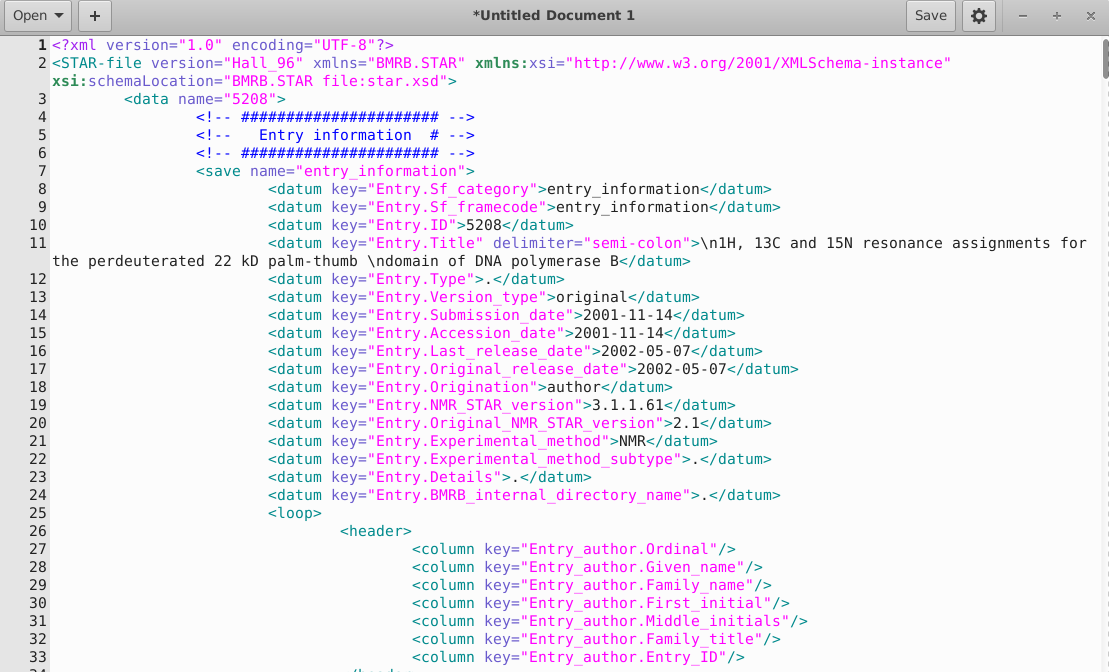
-
STAR XSLT: An extensible stylesheet language transformation has been developed for translating an XML-serialized STAR document back into STAR. Since document order is preserved with both translations, it is easy to inspect the robustness of the translations.
-
STAR syntax highlighting for gedit: The codified STAR grammar was simple to translate into a language definition for the Gnome text editor, gedit. Language definition files are themselves encoded in XML and the STAR language has been upstreamed into the latest gedit release. A screenshot of the syntax highlighting is shown below.
Figure 4: STAR Syntax Highlighting in gedit.
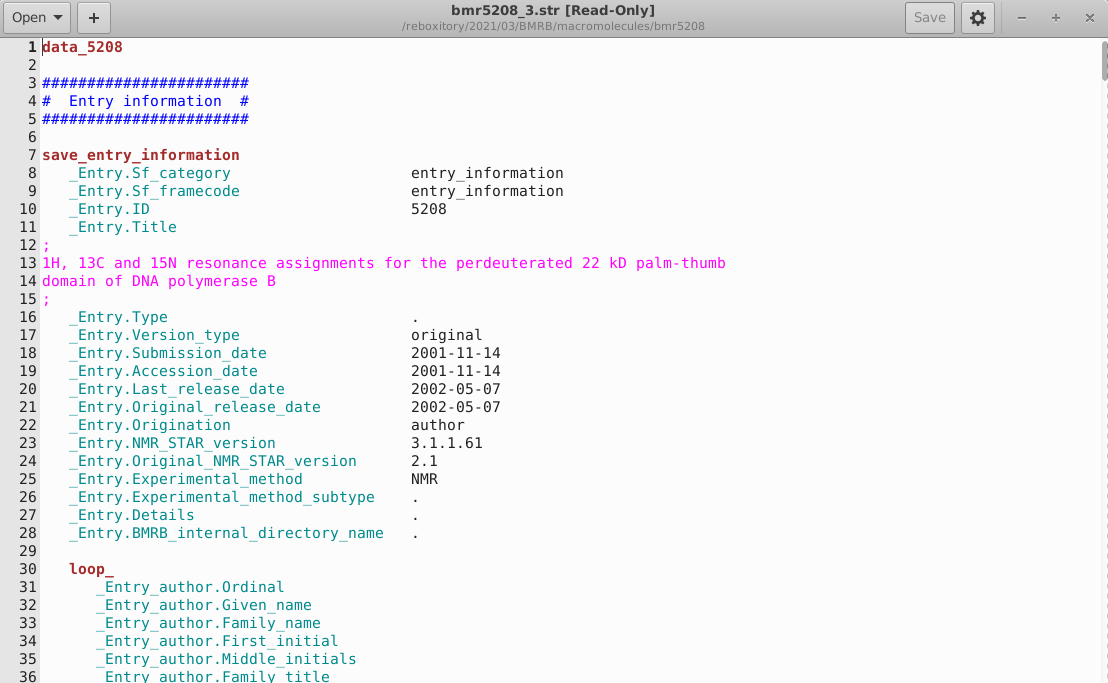
-
NMR-STAR as a Spreadsheet: An extensible stylesheet language transformation has been developed for translating an NMR-STAR document into the Open Document XML Spreadsheet File Format (suffix: .fods). This specification for spreadsheets was chosen because it is an open file format but also importantly because it is a single-file representation of a spreadsheet. This made it straightforward to convert from a single XML file to a single spreadsheet file.
It is worth emphasizing that this conversion is particularly useful in the context of NMR-STAR files due to their restrictions on nested save frames and tables. This makes it straight-forward to cast each datablock as an individual spreadsheet file and each save frame as its own sheet within the spreadsheet file. A screenshot is shown below. (All of the examples use the same underlying BMRB entry, so the reader can compare - at least a portion of - the STAR, XML and spreadsheet representations. Of note, line 4 in the spreadsheet appears to be cut off while in reality it includes a newline symbol and thus the default spreadsheet view does not show it in its entirety. The tabular view of the author names is much more faithful to what a user would expect for such data.)
Figure 5: NMR-STAR as a Spreadsheet.
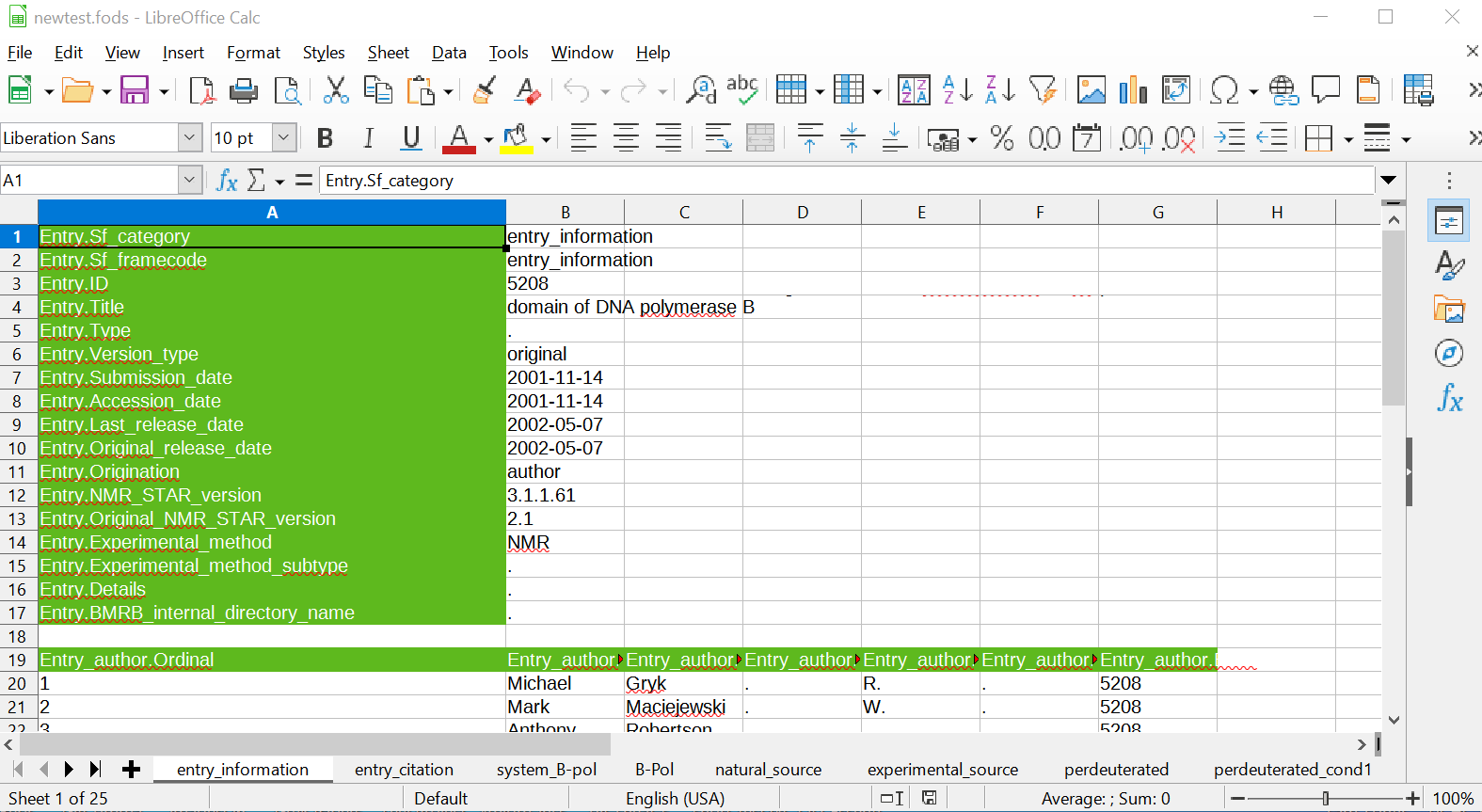
Related Work
The author is not aware of any other efforts to deconstruct the STAR file format as is done in this manuscript or of any other generalized STAR to XML translation tools. There have been tools and schemas developed for converting the specialized STAR data dictionaries into XML. Both the PDB and BMRB have produced XML and RDF schemas for their respective data dictionaries [ Westbrook, et al., 2005; Yokochi, et al., 2016 ]. There are two important distinctions between those efforts and this paper. One, the PDB and BMRB schemas rely on the specific data dictionaries of those domains, particularly how the data items are grouped into categories (save frames). Thus, these schemas and conversion tools cannot translate a generic STAR file to XML but only those which conformed to the supported data dictionary. Two, those domain-specific XML schemas have a specified ordering of elements and cannot preserve document order. Neither of these points are criticisms of the prior work; they are simply distinctions between those works and this one.
Finally, there was also another effort of mapping NMR-STAR to XML using the Document Object Model [ Linge, et al. 1999]. This mapping also relies on the domain specific data dictionary for NMR-STAR.
Acknowledgments
This work was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers GM-111135 and GM-109046.
Appendix A. Example STAR file
data_setA
_location 'New Mexico'
save_observation1
_date 2020-07-01
loop_
_sampleID _height_millimeters
1 6.3
2 2.5
global_
_max_height 6.3
data_setB
_location California
save_observation1
_date 2020-09-15
loop_
_sampleID _height_millimeters
1 9.3
save_observation2
_date 2020-10-15
loop_
_sampleID _height_millimeters
1 9.9
global_
_max_height 9.9Appendix B. Example STAR file illustrating comment ambiguity
# Comments can be at the start of a file
# Should this be file level or a comment on 'data_setA' datablock?
data_setA
# Is this a data block comment or for the following key-value pair
_location 'New Mexico'
save_observation1
_date # what about comments inside pairs?
2020-07-01
loop_
_sampleID _height_millimeters # Or inside loops
1 6.3 # Within columns or rows
2 2.5
# Is this the last comment of the previous block or a comment about the next one?
global_
_max_height 6.3
data_setB
_location California
save_observation1
_date 2020-09-15
loop_
_sampleID _height_millimeters
1 9.3
save_observation2
_date 2020-10-15
loop_
_sampleID _height_millimeters
1 9.9
global_
_max_height 9.9References
[Hall, 1991]
Hall, S.R.
The STAR File: A New Format for Electronic Data Transfer and Archiving
.
J. Chem. Inf. Comput.,
31, 326-333 (1991).
doi:https://doi.org/10.1021/ci00002a020
[Hall, et al., 1991]
Hall, S.R. Allen, F.H., & Brown, I.D.
The Crystallographic Information File (CIF): a New Standard Archive File for Crystallography
.
Acta Cryst.,
A47, 655-685 (1991).
doi:https://doi.org/10.1107/S010876739101067X
[Hall & Spadaccini, 1994]
Hall, S.R. & Spadaccini, N.
The STAR File: Detailed Specifications
.
J. Chem. Inf. Comput.,
34, 505-508 (1994).
doi:https://doi.org/10.1021/ci00019a005
[Hall & Cook, 1995]
Hall, S.R. & Cook, A.P.F.
STAR Dictionary Definition Language: Initial Specification
.
J. Chem. Inf. Comput. Sci.,
35, 819-825. (1995).
doi:https://doi.org/10.1021/ci00027a005
[Allen, et al., 1995]
Allen, F.H., Barnard, J.M., Cook, A.P.F., & Hall, S.R.
The Molecular Information File (MIF): Core Specifications of a
New Standard Format for Chemical Data
.
J. Chem. Inf. Comput. Sci.,
35, 412-427. (1995).
doi:https://doi.org/10.1021/ci00025a009
[Linge, et al. 1999]
Linge, J.P., Nilges, M. & Ehrlich, L.
StarDOM: From STAR format to XML
.
J Biomol NMR,
15, 169–172 (1999).
doi:https://doi.org/10.1023/A:1008347330165
[Westbrook, et al., 2005]
Westbrook, J., Ito, N., Nakamura, H., Henrick, K., & Berman, H.M.
PDBML: the representation of archival macromolecular structure data in XML
.
Bioinformatics,
21, 988-992 (2005).
doi:https://doi.org/10.1093/bioinformatics/bti082
[Ulrich, et al., 2008]
Ulrich, E.L., Akutsu, H., Doreleijers, J.F., Harano, Y., Ioannidis, Y.E.,
Lin, J., Livny, M., Mading, S., Maziuk, D., Miller, Z., Nakatani, E.,
Schulte, C.F., Tolmie, D.E., Wenger, R.K., Yao, H. & Markley, J.L.
BioMagResBank
.
Nucleic Acids Research,
36, D402–D408. (2008).
doi:https://doi.org/10.1093/nar/gkm957
[Spadaccini & Hall, 2012]
Spadaccini, N. & Hall, S.R.
Extensions to the STAR File Syntax
.
J. Chem. Inf. Model.,
52, 1901-1906. (2012).
doi:https://doi.org/10.1021/ci300074v
[Yokochi, et al., 2016]
Yokochi, M., Kobayashi, N., Ulrich, E.L., Kinjo, A.R., Iwata, T., Ioannidis, Y.E.,
Livny, M., Markley, J.L., Nakamura, H., Kojima, C., & Fujiwara, T.
Publication of nuclear magnetic resonance experimental data with
semantic web technology and the application thereof to biomedical
research of proteins
.
J Biomed Semant,
7, 16 (2016).
doi:https://doi.org/10.1186/s13326-016-0057-1
[1] The STAR specification refers to these as data cells, but “cell” is typically used for spreadsheet elements and will not be used here.
[2] The STAR specification uses the term “loop” to refer to tabular data. This is appropriate as the individual data elements are identified by looping through individual columns and rows. However, the term “loop” has such a ubiquitous meaning in programming languages that this paper will mostly refer to them as “tables”. That said, the STAR community has embraced the term “loop” and such the keyword remains and the XML element will be called <loop>.
[3] The STAR loop structure allows for a table to consist of a single row and column. In this case, the loop would be an alternate representation of a key-value pair.
[4] The mmCIF data name “atom_site_anisotrop.U[1][2]” for instance: https://mmcif.wwpdb.org/dictionaries/mmcif_pdbx_v40.dic/Items/_atom_site_anisotrop.U%5b1%5d%5b2%5d.html
[5] The “Core” standard refers to the specification defined in Hall, 1991 and Hall & Spadaccini, 1994. Later work introduces dictionary definition languages which can be used to define schemas [ Hall & Cook, 1995 ] and Spadaccini & Hall, 2012 allows for values to be lists and arrays.