Background
The National Center for Biotechnology Information of the National Library of Medicine originally created the Journal Archiving and Interchange Tag Suite (JATS)JATS01 with the intent of providing a common format in which publishers and archives could exchange journal content.
It was developed in response to a Document Type Definition (DTD) used by the NCBI/NLM PubMed Central project to archive life science journals from a variety of sources. Input and support from Harvard University Libraries, as well as support from The Andrew W. Mellon Foundation and collaboration with Inera, Inc. and Mulberry Technologies, Inc., allowed the scope of the project to be broadened and resulted in the NLM Journal Archiving and Interchange Tag Suite.
JATS is a NISO standard (Z39.96-2012)NISO01 that defines elements and attributes that describe metadata and full content of scholarly journal articles. The Tag Suite is the complete set of elements and attributes described in the standard. Along with these descriptions, the standard includes three article models or Tag Sets: the Journal Archive and Interchange Tag Set, the Journal Publishing Tag Set, and the Article Authoring Tag Set.
The intent of the Tag Suite is to preserve the intellectual content of journals independent of the form in which that content was originally delivered. It enables an archive to capture structural and semantic components of existing material without modeling any particular sequence or textual format.
JATS is the XML model used for all content stored in PMC.PMC01 All content in PMC is free to read, but the XML versions of the truly open access articles are available through the PMC Open Access Subset.PMC02
The Problem
In 2012, an automated software tool—the Open Access Media Importer (OAMI)OAMI1 started using the articles in the PMC Open Access Subset to find audio and video objects that could be loaded to Wikimedia Commons for use on Wikipedia and elsewhere. The OAMI used several JATS elements and attributes including those for licensing, keywords and media types. This use revealed inconsistencies in the XML available from PMC.
Although JATS is a standard and PMC performs some standardization of the submitted XML during ingest, JATS has had to allow for the fine nuances of publishing and the varying requirements of different types of content and different publishers. As a result, publishers use JATS inconsistently, which leads to problems when reusing the content. These inconsistencies affected how the OAMI software could reuse the material. Inconsistencies and ambiguities in license tagging were exceptionally problematic. This required some algorithms to determine whether the content was compatible with reuse on Wikimedia Commons.
For example, in some instances, the license URI specified a “CC-BY” license, but the human-readable text contradicted it, adding an extra non-commercial clause:
<license license-type="open-access"
xlink:href="http://creativecommons.org/licenses/by/2.5/">
<p>Re-use of this article is permitted in accordance with the Creative Commons Deed,
Attribution 2.5, which does not permit commercial exploitation.</p>
</license>In other examples, articles had license information outside of any <license> element:
<permissions>
<copyright-statement> Uosaki et al. This is an open-access article distributed under the
terms of the Creative Commons Attribution License, which permits unrestricted use,
distribution, and reproduction in any medium, provided the original author and
source are credited. </copyright-statement>
<copyright-year>2011</copyright-year>
</permissions>
Other articles were found to have different, contradictory license URIs within the <permissions> element.
Similar problems were found whenever it was attempted to automatically extract metadata related to the article and the accompanying media files. In particular, tagging related to subjects, keywords and captions, the media types of those accompanying files, and other areas.
These problems led to a paperMIET01 that was presented at JATS-Con 2014 and triggered a call to action for the development of best practices for tagging in JATS in a way that improves reusability.BECK01
JATS for Reuse—JATS4R
In June 2014, a group of publishers and aggregators met in Cambridge, UK, to discuss JATS reusability issues. The group formed as "JATS for Reuse" and decided to publish best tagging practices recommendations to improve the reusability of JATS-tagged article content. The meeting resulted in a prioritization list of topics (elements). A websiteJ4R01 and a public mailing listJ4R02 soon followed.
The OAMI work revealed that many other JATS tags are used inconsistently, for example those concerned with mathematical formulas, affiliations and contributor roles.
In January 2015, the group was expanded to include more publishers, representation from online hosts, and also other interested parties such as content processing vendors. The group is open to anyone interested in the creation of content in XML format using the JATS DTD. Current JATS4R endorsing parties are listed on the JATS4R website.
As of this writing, the group has issued tagging recommendations in two areas: Permissions and LicensesJ4R03 and Mathematics.J4R04 When the group formalizes a recommendation, we encode the rules in Schematron. The Schematron files are available from the GitHub repository.J4R05
Tool Details
To facilitate checking instance documents against the JATS4R rules without requiring
journal production editorial staff to set up their own Schematron validation service,
we
have implemented a validation tool hosted at http://jats4r.org/validator/. When
run against an instance document, the tool parses the XML, validates against the correct
NLM or JATS DTD, if one is specified, and then checks it against the JATS4R Schematron
rules, and presents a report to the user. The overall data flow of this tool is depicted
in Figure 1.
Fig. 1: Tool Data Flow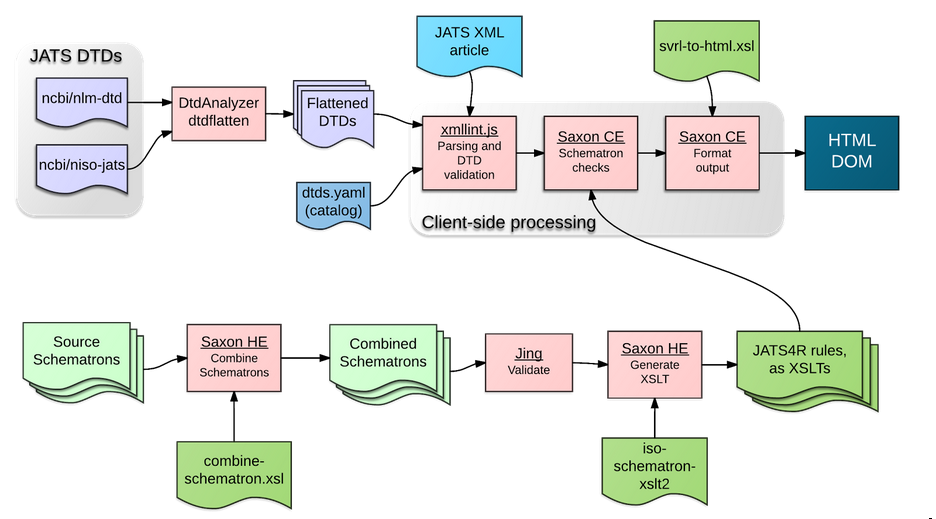
XML Parsing and DTD Validation
Saxon-CESAX01 uses the browser’s native XML parser, which, for most browsers, does not read the DTD (if any) specified in the doctype declaration. This resulted in the tool producing an error if the instance document included entity references defined in the DTD. Therefore, a separate tool was required to parse the documents using the DTD, and resolve those named entity references with their corresponding replacement text. To accomplish this, we have incorporated a JavaScript port of xmllint.
A project on GitHub, xml.js is a port of libxml, including xmllint, to JavaScript, using emscripten. Emscripten is a free tool for compiling C and C++ into optimized JavaScript code. We forked the xml.js project, and made some changes to improve the API.
Within the validator.js module, running on the client, the following code is used to invoke xmllint to parse and validate the instance document:
result = xmllint.validateXML({
xml: [filename, contents],
arguments: ['--loaddtd', '--valid', '--noent', filename],
schemaFiles: [[dtd_filename, dtd_contents]]
});Note that the DTD is passed into the function via the `schemaFiles` argument. The JavaScript implementation of xmllint uses the SYSTEM identifier in the doctype declaration to find the DTD, and validator ensures that the dtd_filename variable matches that SYSTEM identifier.
NLM and JATS DTDs
As alluded to in the previous section, the JavaScript implementation of xmllint does not use OASIS catalogs, and has no way of looking up the correct DTD via the PUBLIC identifier in the doctype declaration. Therefore, before the XML is parsed, the validator uses a regular expression to check for the presence of a doctype declaration. If one is present, it extracts the PUBLIC and SYSTEM identifiers. It uses the PUBLIC identifier to determine which specific NLM or JATS DTD to use to parse the file (there are currently 62 variants). It then also records the SYSTEM identifier as the aforementioned dtd_filename variable, and passes it into xmllint, which dereferences that name to get the DTD contents.
Because this implementation of xmllint has no direct access to a filesystem or to the web, it was important that the DTDs be “flattened”. JATS, and the NLM DTDs before them, are designed modularly; therefore, each variant of the DTD comprises many individual files. We extended the NCBI DtdAnalyzer, adding a new utility dtdflatten, and used it to produce flattened, single-file versions of all 62 variants of the NLM and JATS DTDs. That processing is done offline, when the validator is deployed. We also took the NLM and JATS distributions from the NCBI FTP site, normalized them a bit, and made them available on GitHub, from two repositories, nlm-dtd and niso-jats.
Schematron Representation of JATS4R Recommendations
As described above, the JATS4R recommendations are encoded into schema files in Schematron format. The recommendations are broken down by two categories:
-
level - either errors, warnings, or info
-
topic - math or permissions (currently; more to come)
The Schematron rules for each of the combinations of level and topic are encapsulated in their own files. This is shown schematically in Figure 2. The table in the lower-right shows each of the (six) combinations of level and topic, and each table cell corresponds to a Schematron file.
Fig. 2: Schematron Files
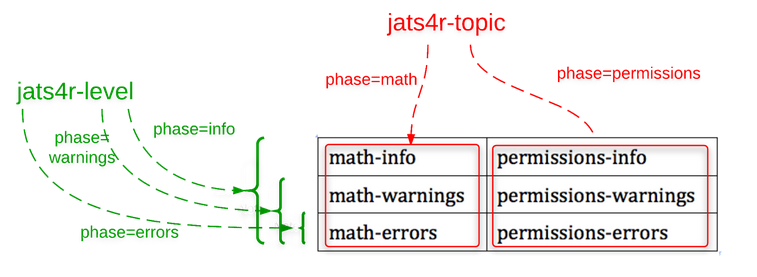
There are two "master" Schematron files which break down the tests in two different ways:
-
jats4r-level.sch - phases for “errors”, “warnings”, and “info”
-
jats4r-topic.sch - phases for “math” and “permissions”
Another "master" Schematron file, which determines conformance or non-conformance of an instance document, is jats4r.sch. This includes all topics, but only the "error” level tests, and is equivalent to using the “errors” phase of jats4r-level.sch.
Schematron Offline Processing
The generated-xsl subdirectory contains XSLT2 files that have been generated from the Schematrons, using the process-schematron.sh script. This script uses conversions available from http://schematron.com.SCH01
This generates separate XSLTs: one for each level and one for each topic. A level-specific XSLT (for example, jats4r-level-errors.xsl) includes the rules for every topic; and conversely, a topic-specific XSLT (for example, jats4r-topic-math.xsl) contains the rules for all of the levels.
The validator has an option box that allows the user to select one of the values for level: errors, warnings, or info. Topic-specific validation using the client-side validator is not available at this time, but could be added easily if there is a demand for it.
Client-side Schematron Validation
The validator runs the instance document through the appropriate XSLT, which generates a report in Schematron Validation Report Language XML (SVRL).
The validator code invokes Saxon, passing the URL of the appropriate XSLT file. The results, in SVRL format, are converted into an HTML report using a separate XSLT transformation.
This is then inserted by Saxon CE into the HTML DOM, and thus presented to the user.
Future work
Validation against Relax NG and/or XSD versions of JATS
This should also be possible without major modifications to the software, since xmllint supports these types of validation.
Port to a server-side validator
Despite our success with implementing this tool on the client, we’ll probably also implement a server-side validator. The main reason for wanting to do this is so that it can be deployed as a web service.
References
[JATS01] JATS Home Page: http://jats.nlm.nih.gov/.
[NISO01] NISO Z39.96-2012: http://www.niso.org/apps/group_public/document.php?document_id=10591.
[PMC01] PubMed Central, http://www.ncbi.nlm.nih.gov/pmc/.
[PMC02] PMC Open Access Subset: http://www.ncbi.nlm.nih.gov/pmc/tools/openftlist/.
[OAMI1] Open Access Media Importer: http://commons.wikimedia.org/wiki/User:Open_Access_Media_Importer_Bot.
[MIET01] Mietchen D, Maloney C, Moskopp ND. Inconsistent XML as a Barrier to Reuse of Open Access Content. In: Journal Article Tag Suite Conference (JATS-Con) Proceedings 2013 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2013. Available from: http://www.ncbi.nlm.nih.gov/books/NBK159964/.
[BECK01] Beck J. Call to Action: http://videocast.nih.gov/summary.asp?Live=13963&start=11980&bhcp=1.
[J4R01] JATS4R website: http://jats4r.org/.
[J4R02] JATS4R mailing list: https://groups.google.com/forum/#!forum/jats4r.
[J4R03] JATS4R Permissions Recommendations: http://jats4r.org/recommendations/permissions.html.
[J4R04] JATS4R Mathematics Recommendations: http://jats4r.org/recommendations/math.html.
[J4R05] JATS4R Validator GitHub Repository: https://github.com/jats4r/validator.
[SAX01] SaxonCE Home Page: http://www.saxonica.com/ce/index.xml.
[NCBI01] Hess D, Maloney C, Hamelers A. DtdAnalyzer: A tool for analyzing and manipulating DTDs. In: Journal Article Tag Suite Conference (JATS-Con) Proceedings 2012 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2012. Available from: http://www.ncbi.nlm.nih.gov/books/NBK100354/.
[SCH01] Schematron to XSL conversion: http://www.schematron.com/tmp/iso-schematron-xslt2.zip.