PubMed Central PMC01 is the U.S. National Institutes of Health's free digital archive of full-text biomedical and life sciences journal literature. Content is stored in XML at the article level. Structures above article (issues, volumes, etc.) are built as collections of articles. The content is displayed dynamically from the archival XML every time that a user retrieves an article. In that respect, every request confirms that the archival copy is still functional.
PMC contains over 2 million article records from over 1,000 titles with 10-15,000 new records being added each month. On average, 600,000-700,000 articles are retrieved by 400,000 unique users each day PMC02.
PubMed Central was started in 1999 to allow free full-text access to journal articles. Participation by journals is voluntary. From the beginning there has always been a requirement that participating journals provide their content to NCBI marked up in some "reasonable" SGML or XML format along with the highest-resolution images available, PDF files (if available), and all supplementary material. Complete details on the PMC's file requirements are available PMC03.
A discussion of what makes a "reasonable" SGML or XML format could be an interesting topic for another paper. For now, "reasonable" means that there is sufficient granularity in the source model to map those elements critical to the understanding of the article (and/or its functioning in the PMC system) from the original article to the appropriate place in the PMC XML model.
The Data Evaluation Process
Journals joining PMC must pass two tests. First, the content must be approved for the NLM collection NLM01. Essentially this is just a check to be sure that the content is "in scope" for a medical library.
Next the journal must go through a technical evaluation to "be sure that the journal can routinely supply files of sufficient quality to generate complete and accurate articles online without the need for human action to correct errors or omissions in the data." PMC03
For the evaluation, a journal supplies a sample set of articles (at least 50). These articles are put through a series of automated and human checks to ensure that the XML is valid and that it accurately represents the article content. There is a set of "Minimum Data Requirements" that must be met before the evaluation proceeds to the more human-intense content accuracy checking PMC04. These minimum criteria are listed briefly below:
-
Each sample package must be complete: all required data files (XML/SGML, PDF if available, image files, supplementary data files) for every article in the package must be present and named correctly.
-
All XML files must conform to an acceptable journal article DTD.
-
All XML/SGML files must parse according to their DTD.
-
Regardless of the XML/SGML DTD used, the following metadata information must be present and tagged with correct values in every sample file:
-
Journal ISSN or other unique Journal ID
-
Journal Publisher
-
Copyright statement (if applicable)
-
License statement (if applicable)
-
Volume number
-
Issue number (if applicable)
-
Pagination/article sequence number
-
Issue-based or Article-based publication dates. Articles submitted to PMC must contain publication dates that accurately reflect the journal’s publication model.
-
-
All image files for figures must be legible, and submitted in high-resolution TIFF or EPS format, according to the PMC Image File Requirements.
These seem like simple and obvious things - xml files must be valid - but the minimum data requirements have greatly reduced the amount of rework that the PMC Data Evaluation group has to do. Certainly it helps to be explicit about even the most obvious of things.
PMC Processing Model
The PMC processing model is diagrammed in Fig. 1. For each article, we
receive a set of files that includes the text in SGML or XML, the highest resolution
figures
available, a PDF file if one has been created for the article, and any supplementary
material
or supporting data. The text is converted to the current version of the NLM Archiving
and
Interchange DTD JATS01, and the images are converted
to a web-friendly format. The source SGML or XML, original images, supplementary data
files,
PDFs, and NLM XML files are stored in the archive. Articles are rendered online using
the NLM
XML, PDFs, supplementary data files, and the web-friendly images.
Fig. 1: PMC Processing Model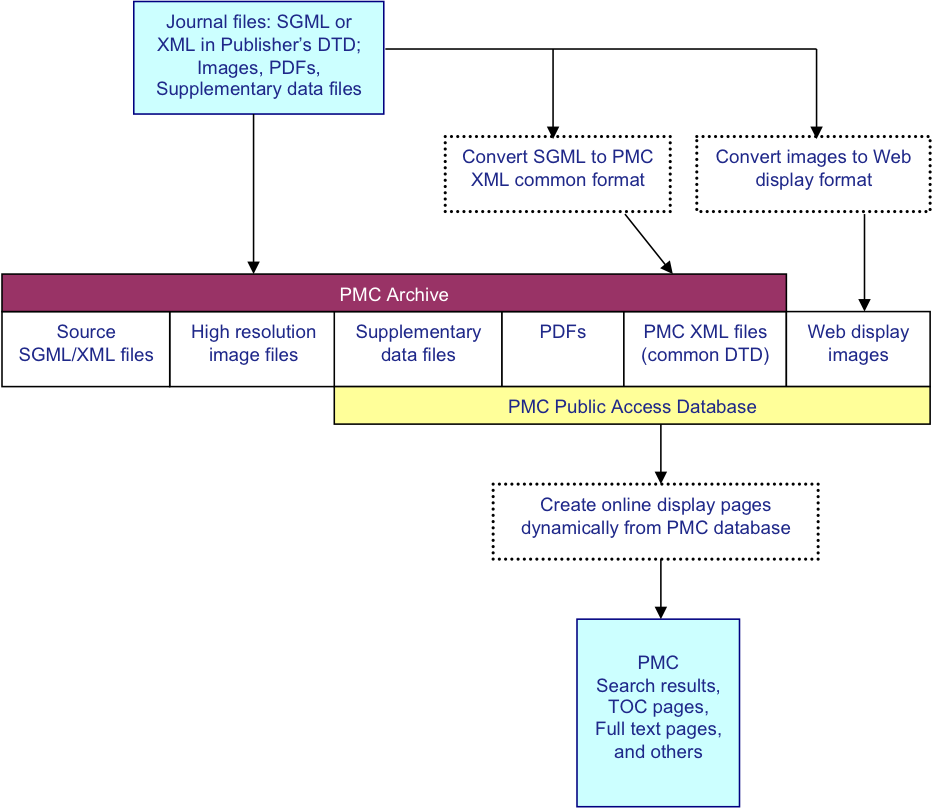
This processing model seems obvious now, but this is not how the content was always
handled. The early PMC processing model is shown in Fig. 2.
Fig. 2: Original PMC Processing Model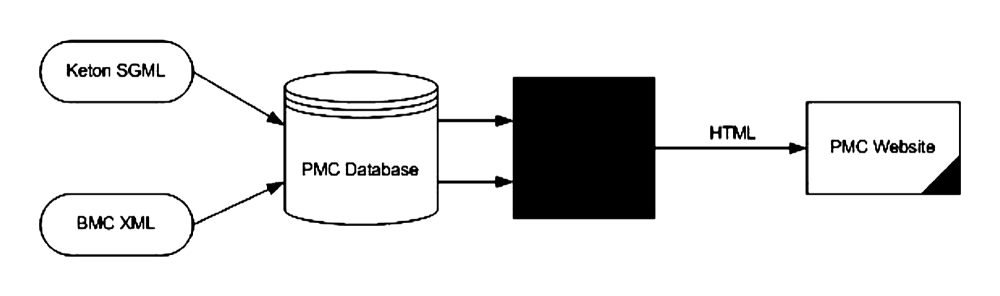
As we started getting more and more submitters, it became obvious that we needed to
take
the processing hit of conversion to a single text format up front and allow the database
and
rendering system concentrate on only one type of XML (Fig. 3). We created the
pmc-1.dtd PMC05, which was based on the first
two submission DTDs that we had to PMC: keton.dtd, an SGML DTD somehow descended from
ISO12083, and the BioMed Central article.dtd, an XML DTD that was very similar.
Fig. 3: Updated PMC Processing Model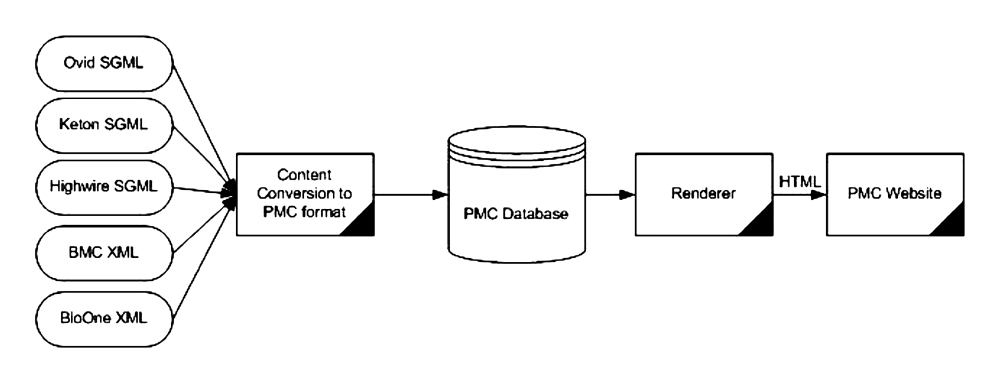
The pmc-1.dtd was written to simplify access to full-text articles online. It was a small and simple DTD that soon showed its limitations as we started to convert more submission formats into it. It grew quickly, and quickly grew unwieldy. At this point, work began on a new DTD that became the NLM Archiving and Interchange Tag Suite Beck01.
XML Philosophy
There are a number of things in our XML Philosophy, which may at first seem contradictory, that have lead to the success of PMC and the manageability of the article ingest work. In general, we have found a balance between strictness and flexibility that allows us to keep a certain amount of control but not be too restrictive on our submitters.
Some Strict Things
The first thing we are strict about is that we do a complete review of any new DTD in which content is being submitted, as described above. We do not take articles in HTML. We also do a complete review of sample articles for each new journal to be sure that the content provider is able to provide content that is structurally and semantically correct.
Another thing we are strict about is that all content must be valid according to the DTD in which it was submitted - not just during data evaluation but in the ongoing production process as well. This seems like an obvious one, but there was a surprising amount of controversy about this in the early days of PMC, and we still get invalid files. Problems usually arise now because the submitter has made a DTD change (as simple as adding a new character entity to the DTD) without telling us or sending an updated DTD.
Also, we do not fix text; all content changes must be made by the submitter, and the content must be resubmitted.
Some Flexible Things
Some things we are not so strict about, which reduces some of the burden on our submitters. First, we don't require all content to be in our format or to follow our tagging rules. We don't force updates of content to latest DTD version, and we generally follow journal style where it does not interfere with our systems.
The DTD
We use the NLM Archiving and Interchange DTD as the format for all articles loaded to the PMC database. This model was created specifically for archiving article content. It was designed to be an "easy target to hit" when transforming content from the over 40 different input models that we receive content in. Currently we are writing content into version 3.0.
We do not migrate all content to each new version of the NLM DTD when it comes out. The system is robust enough to handle content from version 1.0 of the DTD through version 3.0, so we are not constantly churning the data.
We manage all of the versions of the DTD with an XML Catalog, which we also use to manage all of the input DTDs (SGML and XML). We maintain all mappings of PUBLIC and SYSTEM IDs for any DTD that we use in the XML catalog on our Linux machines and then create other catalogs from it each time it is updated. We create an SGML Catalog for the SGML tools that we use; a single "Oxygen" catalog that everyone on the team can use over the network with the XML editor; and a copy of the catalog that refers to http-based copies of the DTDs for PMC International sites. The XML Catalog is an essential piece of the PMC system.
PMC Tagging Style
Next, we've defined a set of rules for objects within articles that is more restrictive than the DTD. This allows us to have normalized structures (figures, tables, contributors) in articles for ease of processing and rendering. We call these rules the PMC Tagging Style, and all articles must "pass style" before being loaded to the database. They are documented in the PMC Tagging Guidelines PMC06.
(Re)Usability of XML
Finally, our XML must be useable by others. The NLM XML that we create from whatever was submitted to us is always available to the submitting publisher (the content owner), and a subset of the articles that are Open Access are available to anyone for download through the PMC Open Archives Service PMC07. This keeps us honest. We can't allow ourselves to take shortcuts with the data. All articles must be valid according to the public NLM DTD version that they reference, and we only use Processing Instructions for instructions about processing.
PMC Tools
Another thing that has been helpful is that we share tools we've built with our submitters. The more help we can give to people sending us content, the cleaner the content is, and the easier it is for us to process. Reducing or eliminating correction cycles greatly reduces cost and stress on the PMC team.
-
PMC Tagging Guidelines PMC06 - This is a document that describes the PMC Tagging Style mentioned above. We first created the guidelines as an internal document that we would use to keep track of any tagging rules we set up to keep our XML as normal as possible. As we got more and more questions from providers about how structures should be tagged, we decided to share our internal document.
The DTD allows many ways to tag certain structures, but we do have a preference for many of them, and sharing them has been helpful both for us and for PMC submitters. While we don't require content that is submitted to PMC be "to style", we certainly aren't going to turn it away.
-
PMC StyleChecker PMC08 - This is a tool that applies the rules defined in the Tagging Guidelines. It is an XSL transform that reviews each article and reports what it finds. This is the same transform that we have inline in our production process after validation against the DTD to confirm that the structures withing the articles we are loading into the database meet our standards for tagging.
It is available as an online tool that users can upload one article at a time or as a set of stylesheets that data providers can build into their workflow PMC09. ().
-
Online SGML Validator PMC10 - This is a tool that validates SGML against the SGML DTDs that we have in the PMC system as long as the PUBLIC or SYSTEM ID is recognized in our SGML Catalog file. That is, as long as we have set up the particular SGML DTD for ingest in our system.
-
Online XML Validator PMC11 - This is a tool that validates any XML file whose SYSTEM ID is resolvable on the system or whose PUBLIC or SYSTEM ID is mapped in our XML Catalog file.
-
PMC Article Previewer PMC12 - This is a tool that runs an article through a PMC ingest workflow. First the SGML or XML is validated against the DTD per the rules given above for the SGML and XML validators. Then, if we recognize the DTD and have a conversion built for it, the article runs through the XSL transform to normalized PMC XML. This output is validated against the output DTD and is checked with the StyleChecker. If successfull at all of these steps, the article is displayed on a page in PMC display format so that the publisher can preview the article. Also, he has a good idea that his article will be ingested into PMC with no problems.
The Article Previewer requires users to login with a "My NCBI" account NCBI01 because the content stays on the database until the user removes it.
Our People
Finally, there has to be some acknowledgment of the people who work on the project. We have a fantastic set of programmers that builds us some very nice software, and we have a good group of Content Managers who marshall the content through the workflow and deal with the exceptions and communicate with the publishers. But, we have a group in the middle that writes and maintains the XSL transforms.
It is not easy to find the right people for this group. Our document conversion writers need to have the strong attention to detail, unending curiosity, and a desire to always find a better way to do things of a good programmer. But, they also need to live in a world where mixed-content does not frightent them. Because we are writing transforms to maintain the meaning and intend of the origingal article, we slip out of the black-and-white prgrammer world into a world of grays. We need to represent the article that was submitted to the archive - and not necessarily the file that was submitted.
A successful PMC document conversion writer lives with a foot in both camps: the world of the wants and desires of the content owners and the rule-driven world of people who build and run large systems. This is not something that is unique to PubMed Central by any means, but it does surprise some people that our group is made up of people who studied English, Art, and Library Sciences rather than Systems Design, COBOL or The DOM.
Because we work exclusively with documents and document XML, I have found that it is much easier to start with a (technically oriented) person with a publishing or printing background and teach them how to write a transform than to take a brilliant programmer and teach them how to think about documents.
Some Challenges: When Print Goes Away
There are still a few things that give us problems in PMC. First, because of the rapid changes in article publishing since the invention of the Internet, journal publishing models have started to change. A business that had a pretty stable model of articles in issues that printed and mailed on a periodic schedule has been shifting to publishing on a continuous basis - with articles made available online as soon as they are ready and then collected into print issues after some time (or not collected and printed at all).
Early PMC was created based on the issue model, and we have had to be flexible to handle these changes.
One thing that we have done to remain flexible here to allow us to deal with this continuous publication model is to be very strict about publication dates in articles. Depending on the publication model of a journal, an article may have one publication date (for traditional print-based journals) or an article-level publication date and a collection-level publication date (for articles that are published online and then collected into a print or online issue. volume, or collection. See the essay on <pub-date> in the Tagging Guidelines PMC06.
The rise of the internet has also brought about another "Continuous publication" idea, and that is continuous publication of the same document - or updates to an article. We have not seen too much of this in PMC yet, but we have been preparing ourselves. Each article in the PMC database is assigned an ArticleID and an ArticleInstanceID. Any new version of an article creates a new ArticleInstance, and all ArticleInstances for a given article have the same ArticleID. This allows us to store multiple versions of articles, retrieve the latest (by ArticleID), and to retrieve any version (by ArticleInstanceID). Fortunately, how the Journal Publishers will handle will need to be decided by the Journal Publishers.
Conclusion
The success we have had at PMC has really been due to a combination of factors including our strictness to upholding the XML models that we use, our flexibility (to a point) that we have with our content providers, the tools that we've put together to make our jobs and the jobs of our content providers easier, and teh fine people we have working on the team. To be fair, it is also nice to have the resources of the National Center for Biotechnology Information and the National Library of Medicine behind us as well as the support of the NIH in general, who have declared that copies of all peer-reviewed articles that report on NIH-funded research must be deposited to PubMed Central and made available within 12 months of the publication date Beck02. I have to include that just to be fair.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Library of Medicine, National Center for Biotechnology Information.
References
[PMC01] PubMed Central, http://www.ncbi.nlm.nih.gov/pmc/.
[PMC02] PubMed Central National Avisory Committee. (2009) Minutes of the PubMed Central National Avisory Committee Meeting, June 15, 2009. National Institutues of Health, Bethesda, MD.
[PMC03] How to Join PMC, http://www.ncbi.nlm.nih.gov/pmc/about/pubinfo.html.
[NLM01] NLM Collection Development and Acquisitions, http://www.nlm.nih.gov/tsd/acquisions/mainpage.html.
[PMC04] Minimum Criteria for PMC Data Evaluation Submissions, http://www.ncbi.nlm.nih.gov/pmc/pmcdoc/mindatareq.pdf.
[JATS01] NLM Archving and Interchange DTD, http://dtd.nlm.nih.gov/archiving/.
[PMC05] pmc-1.dtd, http://www.ncbi.nlm.nih.gov/pmc/pmcdoc/dtd/pmc-1.dtd.
[Beck01] Beck and Lapeyre (2003) New Public Domain Journal Archiving and Interchange DTDs. XML 2003. http://www.idealliance.org/papers/dx_xml03/papers/04-01-02/04-01-02.html.
[PMC06] PMC Tagging Guidelines, http://www.ncbi.nlm.nih.gov/pmc/pmcdoc/tagging-guidelines/article/style.html.
[PMC07] PMC Open Archive Interface, http://www.ncbi.nlm.nih.gov/pmc/about/oai.html.
[PMC08] PMC Online StyleChecker, http://www.pubmedcentral.nih.gov/utils/style_checker/stylechecker.cgi.
[PMC09] PMC Downloadable StyleChecker, http://www.ncbi.nlm.nih.gov/pmc/pmcdoc/tagging-guidelines/stylechecker/stylecheck-README.html.
[PMC10] PMC SGML Validator, http://www.pubmedcentral.nih.gov/utils/validate/sgmlcheck.cgi.
[PMC11] PMC XML Validator, http://www.pubmedcentral.nih.gov/utils/validate/xmlcheck.cgi.
[PMC12] PMC Article Previewer, http://www.pubmedcentral.nih.gov/utils/pv/.
[NCBI01] My NCBI, http://www.ncbi.nlm.nih.gov/sites/myncbi/.
[Beck02] Beck, Jeff. “How XML made the NIH "Policy on Enhancing Public Access to Archived Publications Resulting from NIH-Funded Research" possible.” In Proceedings of Extreme Markup Languages 2005. http://conferences.idealliance.org/extreme/html/2005/Beck01/EML2005Beck01.html